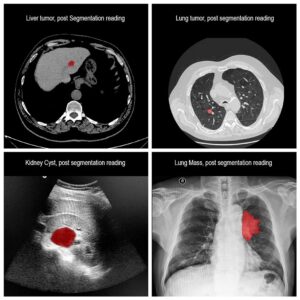
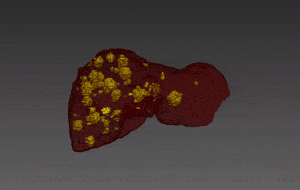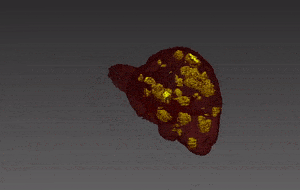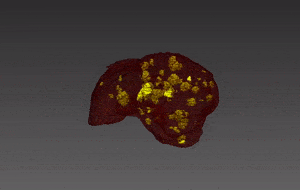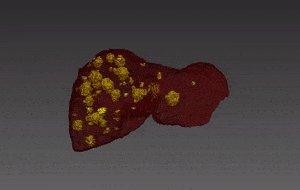
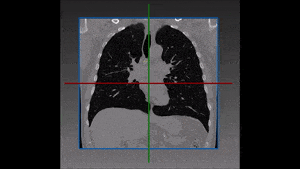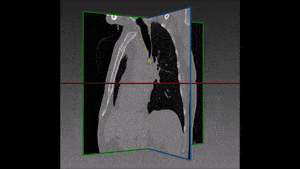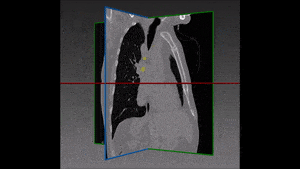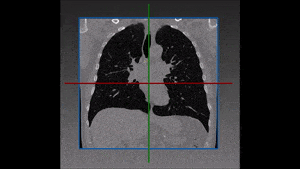
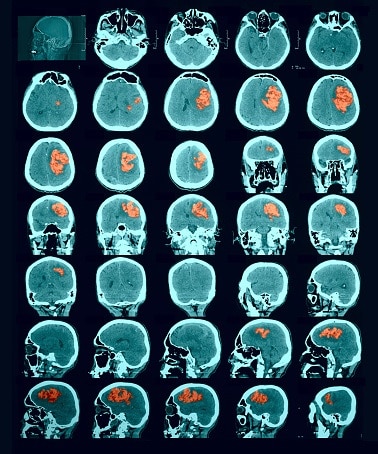
Segmentation of lesions in images, such as those obtained from MRI, ultrasound, CT etc, can be viewed as classifying pixels (or voxels, in the 3-D case) of the image into one or more classes. In the general case, several classes are used, indicating the membership, or probability of each pixel to belong to the lesion region. In the simplified version, a binary classifier is used to make decisions regarding the membership of each pixel to lesion.
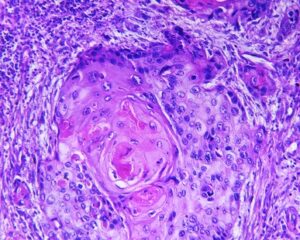
In practice, lesions appearance vary significantly in shape, size, color and texture, and can appear differently when visualized by different modalities. Constructing deterministic algorithms to segment lesions can likely be successful only in limited, well studied cases. To overcome such variability in lesion appearance, statistical classifiers are used. These classifiers can be taught to mimic expert decision making based on image features values. The set of feature values is determined beforehand and can take up different forms and include both spatial and chromatic information for each pixel or region. In addition, higher order relationship between pixels or image regions can be used as features.
One popular supervised classification method is the random forest classification. Random forest are trained classifiers, used to delineate the boundaries between points in feature space belonging to different classes, with the goal of maximizing the margin between classes (similar to Support Vector Machine classifier). The random forest is composed of an ensemble of decision trees, trained to take input values (feature vector) and assign membership value to either the lesion or background group. It is worth noting that, similar to histogram or frequency analysis of the image, classification based on feature values can significantly reduce the classification space.
Using a single decision tree turns out to be a very bad classifier. However, the majority vote by an ensemble of trees performs significantly better in segmentation.
Training a random decision tree involves determining decision rules in each node of the tree. Decisions, most commonly binary decisions, are merely quantitative tests on the input feature value. A simple test can be setting a threshold, by which input feature values is passed to the right child node is feature exceeds the threshold, and to the left otherwise. This is how information ‘travels’ down the tree, subjected to a test in each node. The threshold value, or the quantitative test, can be determined from a pool of tests by choosing the one which maximizes the information gain. Information gain can be computed, by the data entropy. Data is passed from the tree top to its children until reaching the leaf (end node), where a decision is made. Averaging the possible output in all leafs, is one way of obtaining the decision of the tree.
In practice, to set the threshold value in each node, we pass the information (values of the parameters in the feature space) through the tree and then keep the rules (or node) which maximizes the information gain. The fact that training is supervised allows us to keep in only nodes (decision rules) which maximize the success of classification.
To construct each tree, we present it with a different bootstrap subset of the training data (feature values) chosen at random. For this reason, the decisions produced by each tree will be slightly different. Since trees are uncorrelated, the overall decision in random forest can be shown to be consistent by using (for example) majority vote of trees with different structure (assuming enough trees are used).
Both technical and conceptual challenges pile up fast when constructing such classifiers for complex tasks like lesion segmentation. These challenges are regularly dealt with success by engineers at RSIP Vision. Our specialty is tailoring computer vision and machine learning algorithms for complex industrial and medical applications. The expertise cultivated at RSIP Vision has allowed us to construct cutting edge solutions for countless clients worldwide.
One popular supervised classification method is the random forest classification. Random forest are trained classifiers, used to delineate the boundaries between points in feature space belonging to different classes, with the goal of maximizing the margin between classes (similar to Support Vector Machine classifier). The random forest is composed of an ensemble of decision trees, trained to take input values (feature vector) and assign membership value to either the lesion or background group. It is worth noting that, similar to histogram or frequency analysis of the image, classification based on feature values can significantly reduce the classification space.
Using a single decision tree turns out to be a very bad classifier. However, the majority vote by an ensemble of trees performs significantly better in segmentation.
Training a random decision tree involves determining decision rules in each node of the tree. Decisions, most commonly binary decisions, are merely quantitative tests on the input feature value. A simple test can be setting a threshold, by which input feature values is passed to the right child node is feature exceeds the threshold, and to the left otherwise. This is how information ‘travels’ down the tree, subjected to a test in each node. The threshold value, or the quantitative test, can be determined from a pool of tests by choosing the one which maximizes the information gain. Information gain can be computed, by the data entropy. Data is passed from the tree top to its children until reaching the leaf (end node), where a decision is made. Averaging the possible output in all leafs, is one way of obtaining the decision of the tree.
In practice, to set the threshold value in each node, we pass the information (values of the parameters in the feature space) through the tree and then keep the rules (or node) which maximizes the information gain. The fact that training is supervised allows us to keep in only nodes (decision rules) which maximize the success of classification.
To construct each tree, we present it with a different bootstrap subset of the training data (feature values) chosen at random. For this reason, the decisions produced by each tree will be slightly different. Since trees are uncorrelated, the overall decision in random forest can be shown to be consistent by using (for example) majority vote of trees with different structure (assuming enough trees are used).
Both technical and conceptual challenges pile up fast when constructing such classifiers for complex tasks like lesion segmentation. These challenges are regularly dealt with success by engineers at RSIP Vision. Our specialty is tailoring computer vision and machine learning algorithms for complex industrial and medical applications. The expertise cultivated at RSIP Vision has allowed us to construct cutting edge solutions for countless clients worldwide.

 Medical segmentation
Medical segmentation