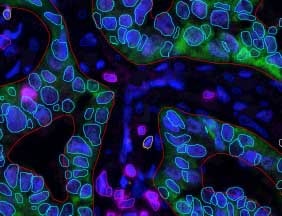
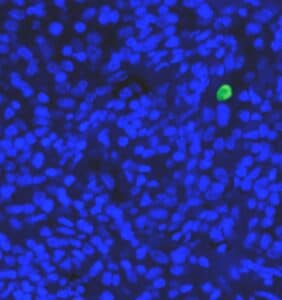
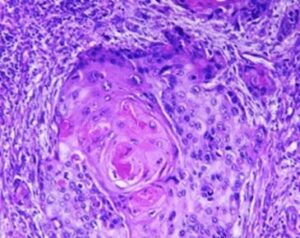
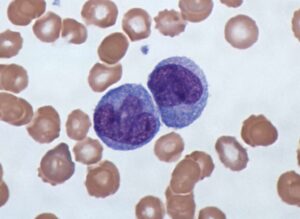
Several prognostic factors indicate the state and progression of breast cancer. Grading of the state of progression and spreading of cancer enables to start the suggested treatment. In solid tumors, like breast cancer, one such prognostic factor is the mitotic figure, i.e. a cell whose chromosomes appear as dark stains. In a histological sample taken from patients, the fraction of breast tissue cells undergoing replication (mitosis) is used to grade the cancer according to scales defined by the oncologist, who in turn suggests the right avenue of treatment.
Abnormal nuclei deformation is the key feature disclosing the mitotic state of a cell. Even in a stained histological sample, in which all nuclei are painted (e.g. blue), extracting the mitotic figure is not a trivial task. Substantial histological training is required to distinguish mitotic from non-mitotic nuclei due to their high similarity. Furthermore, heterogeneity of tissue within the histological sample makes it difficult to assess the actual mitotic figure belonging to the breast tissue.
Detection, recognition, and classification can be performed several orders of magnitude faster using automatic computational autonomous tools. The great success of deep neural networks lies in that they help distinguish complex patterns in images, which makes them an attractive option for mitotic figure estimation from histological samples.
Briefly, deep neural networks extend the traditional neural network by adding several types of neuronal layers. Just as in traditional neural network, input data is passed between nodes (neurons), whereas in each neuron an activation function processes the input and transfers the output to the neurons connected to it. The process continues to give output in several nodes at its end, representing a class probability each.
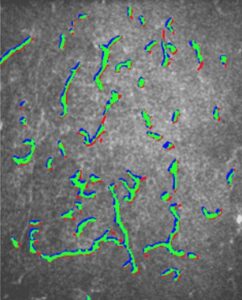
Dealing with histological RGB images, a deep neural network needs to classify each pixel (or region) into mitotic or non-mitotic, making it essentially a binary classifier. It is an art to find the correct network architecture that will perform best, given the training data. Since images are used as input, deep neural networks are equipped with convolution layers, which basically apply a filter to a 2D image.
Max pooling layers are used to select the best features for the recognition of mitotic nuclei. These layer cannot be trained and perform as constant filters to the data. For example, if an activation function operates on non-overlapping squares of the image, the max-pooling layer will give as an output the maximal value of the activation function over all squares.
All information passed through the network architecture eventually end up in two nodes, representing the probability of an input information to belong to one of either mitotic or non-mitotic class. To be able to utilize deep neural networks for complex classification procedure, a great expertise is required on the developers end. The expert’s knowledge, in this case the histologist’s, needs to be transferred into training data which is used for the adjustment of weights in nodes of the network. Furthermore, the actual architecture – with the right choice of layers and the inter-connectivity – is of crucial importance.
At RSIP Vision we hold such expertise. Our engineers have long been dealing with classification algorithms in the biological domain. Neural networks are routinely tailor-made at RSIP Vision to solve classification problems. See our portfolio to learn how RSIP Vision can employ deep neural networks to the benefit of your projects.

 Microscopy
Microscopy