Medical imaging and medical image data analysis are rapidly growing fields. The increasing amounts of available data due to advances and ubiquity of imaging technologies give rise to new medical applications and to new requirements in existing applications, and lead to an increasing demand for new and better data analysis methods and algorithms. One such example is minimal-intervention operations. Such surgeries are on the one hand possible due to advanced real-time imaging methods, but on the other hand, require precise automatic tracking of surgical tools – thus posing new algorithmic challenges to the field.
In recent years, there have been significant advances in the field of neural-network based machine learning methods. With these advances, Deep Learning (DL) methodologies have also entered the medical field, and have become a prominent framework for medical imaging data analysis; the term Deep Learning refers to the large number of layers of modern neural networks, often called the “depth” of the network. For example, in MICCAI 2017 – one of the central conferences for medical imaging research – more than one out of every four papers was directly concerned with deep learning. Deep learning models are able to learn complex functions, are suitable for dealing with the large amounts of data in the field, and have proven to be highly effective and flexible in many medical imaging tasks.
The current paper aims at reviewing the recent advances in applications and research of deep learning in medical imaging. Since both the field of medical imaging and the field of deep learning are advancing fast, here we focus only on the most recent developments in the field, and we refer the reader to [43] for a review of other works prior to 2017. There are many sub-fields to medical imaging, and it is possible to review the works according to many categories: according to the medical branch, imaging modality, the algorithmic task at hand (e.g., classification, segmentation, super resolution, image registration), or the deep learning solution technique that was utilized or developed for the problem.
In order to both give a wide picture about the state-of-the-art of Deep Learning in medical imaging and make this review useful as a map for researchers and practitioners in the field, we chose to survey works according to several medical branches in the following sections, and to use this introduction to give a breadth-wise overview of central themes that arise – from the algorithmic perspective – across the different medical fields and imaging modalities, and across different works from the recent year. In this series we discuss deep learning in medical imaging in the fields of Brain Imaging, Ophthalmology, Pulmonology and Cardiology. The central themes we discuss next are advances in the use of convolutional neural networks, the increasing use of long-short-term-memory networks for various medical tasks, algorithms involving learning with weak supervision, hybrid or indirect uses of Deep Learning, combination of prior knowledge with neural networks, and multi-modality Deep Learning methods.
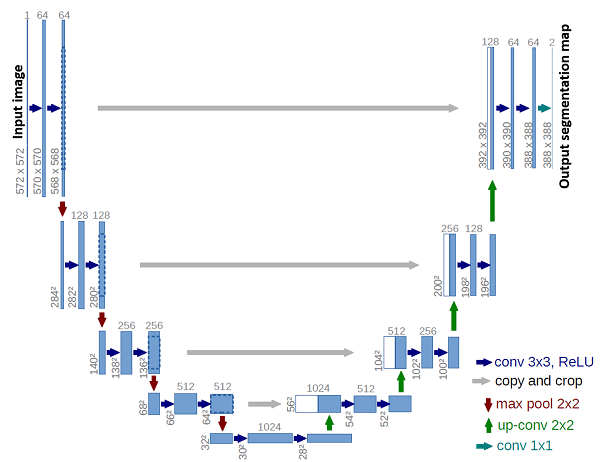
An illustration of The U-Net encoder-decoder architecture [53].
The left branch is the encoder part of the network
and the right branch is the decoder. The gray arrows represent
skip connections at four compression and processing levels.
.
Convolutional Neural Networks: Convolutional Neural Networks (CNNs) play a key role in modern image data analysis and computer vision. CNNs are capable of extracting informative, translation invariant features from image data or volumetric data, and have shown usefulness for many medical imaging purposes and imaging modalities. A prominent CNN macro-architecture is the encoder-decoder structure, where input data is compressed to a dense representation through several layers which comprise the encoder part of the network, and then information is re-interpreted in the decoder layers, usually to output a pixel-wise classification or regression information for object detection or image segmentation. Two well-known and effective network architectures of this type are U-Net [53] (see figure above) and SegNet [8], and many recent advances in the use of CNNs include adjustments and refinements of this idea, and suggest task-specific network architecture modifications. Recent advances in this vein include CT and MR volume registration [62, 59], registration with motion correction [29, 58, 35], tissue image segmentation [27, 26, 69, 14, 45, 72, 57, 75, 63, 6, 13, 38, 73, 55, 12, 17], medical instruments segmentation in images [40, 51, 74, 42, 4], super resolution and image enhancement [61, 47, 9], and diagnosis assistance or pathology classification [7, 68, 5, 76].
Long Short Term Memory Networks: Long Short Term Memory Network (LSTM) is a recurrent neural network architecture that was originally designed to deal with the problem of vanishing gradients during the training of recurrent networks on long temporal sequences using gradient descent methods [28]. The LSTM structure includes a gated state-cell which is selectively and linearly manipulated throughout the processing of a temporal sequence. This solution of the original error gradient problem enables the network to learn long range temporal structure, as the state-cell effectively serves as a memory, enabling the network to keep track of previous events. Due to their ability to learn structural data and model long-range temporal relations, LSTMs are commonly used in natural language processing. However, recent works show that LSTMs also prove useful for medical imaging tasks when combined with features learned by convolutional neural networks: either with explicitly temporal data [67, 30, 1], or by modeling spatial data as a sequence [50, 65, 66]. Another interesting application was suggested in [3] who use LSTMs for real-time prediction of the time remaining for a surgery, for operation room time optimization in hospitals.
Weak Supervision: Obtaining annotated medical data requires many hours of annotation by medical experts. For many tasks such as image segmentation, or 3D volume registration, the annotation requires pixel-level attention, and for tasks involving video data a frame-level annotation is needed; both are time consuming. Hence, many medical imaging challenges suffer from the problem of large datasets, but with only small numbers of annotated examples – a problem that practitioners and researchers in the field are too well acquainted with. This problem forms a stringent limitation on development of new applications where existing annotated data is not available.
Several recent works suggest solutions for this problem, using different approaches: [56, 10, 11] combine unsupervised learning techniques with supervised training on a small labeled dataset; [54, 62] use automatically generated model-based labels for data augmentation, and show that networks trained on a combination of human-labeled and model-labeled data outperform the original model; and [20, 21] show how to train networks for segmentation using only high-level annotation, rather than pixel-wise labels. For a specific review on semi-supervised learning in medical imaging see [15] (pre-print version). Other approaches deal with sparsely annotated datasets in a non-direct manner. [25] study the optimal ways to perform transfer learning for MRI segmentation, and other works (as we will discuss next) combine deep learning either with classic algorithms, or with prior information and modeling assumptions.
Hybrid Models and Indirect Deep Learning: Another prominent direction in recent studies is the design of hybrid models which utilize Deep Learning methods in a non-direct manner, as a part of another algorithm – either combining them with other machine learning methods, or with classic tools in specific tasks. [39] use Graph-CNNs in a non-direct manner to learn functional relations between brain regions. [71] use a CNN to learn a coarse solution for brain-fiber orientation mapping, and use this solution as a regularizing term for a classic regression solution (see Section 2). [36] combine hand-crafted and CNN features with classic machine learning for prostate lesion classification. [49] perform cardiac motion tracking in temporal echocardiography data using a model of graph flows; they learn the graph edges with a Siamese CNN and solve the flow over this graph with anatomical model based methods (see Section ??). [18] input CNN features together with patient information to a random forest algorithm for cyst classification.
Deep Learning with Prior Information: Recent papers discuss and suggest methods for incorporating prior knowledge to deep learning. Prior knowledge can be, for example, known anatomical constraints, statistical knowledge, or knowledge obtained by other methods (before training a neural network). The inclusion of prior information in the training of a deep learning model is not a trivial task, since the representation of the data by the network is not directly controllable, and generally not easily interpretable before the final output of the network (the network acts as a black box). Hence, creative solutions are required for taking advantage of both prior knowledge and deep learning tools.
Recent works propose such solutions in different contexts. [52] and [48] use auto-encoders to learn shape priors and combine them with CNNs as regularizers, for kidney ultrasound segmentation, for cardiac MRI and ultrasound segmentation, and for super-resolution. Though both their methods incorporate priors obtained from unsupervised learning, and not from direct anatomical knowledge, the methods may be applicable to other forms of shape priors as well. In [2], anatomical information is encoded into the network input for bone landmark modeling. They use geodesic patch extraction to obtain local shape descriptors, and then use these as input data to train a Siamese CNN on classification of correspondence and non-correspondence of shape patches. [46] combine CNNs with principle component analysis to achieve more stable results.
Multi-Modality: Medical data analysis tasks and real time medical operation imaging tasks often involve different types of data about the same patient. Several recent works address the question of how to integrate these data from different modalities for the performance of a single task: [44] design a multi-branch CNN that combines data from different brain patches, as well as patient personal information, for brain disease diagnosis; [22] perform brain tumor segmentation in MRI data by effectively splitting a CNN to several channels for different MRI modalities, with sparse interaction between the channels; and [34] perform multi-input MR image synthesis (see section on brain imaging). [24] train a CNN for skin disease recognition with inputs of both macroscopic mole images and the corresponding dermatoscopy imaging. [64] demonstrate how to use ensemble learning for integrating different inputs for benign-malignant lung nodule classification, and [16] suggest a creative solution for combining textual information about the patient in CNNs with fundus imaging for diabetic retinopathy detection.
The above summary is of course not comprehensive, and there are many other works worth mentioning that we have not referred to. As the field of medical imaging is a very active field, both in the academy and industry, new developments even come up as we write (and as you read) this text. However, it is our hope that this review conveys a good description of the current trends and latest advancements in the field, and that the coming sections will provide a useful map of references for the most important works and methodologies in the specific medical branches: the fields of Brain Imaging, Ophthalmology, Pulmonology and Cardiology.
Deep Learning in Medical Imaging – References
(*) Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) Medical Image Computing and Computer-Assisted Intervention MICCAI 2017. Springer International Publishing, Cham (2017)
[1] Abdi, A.H., Luong, C., Tsang, T., Jue, J., Gin, K., Yeung, D., Hawley, D., Rohling, R., Abolmaesumi, P.: Quality assessment of echocardiographic cine using recurrent neural networks: Feasibility on five standard view planes. Printed in (*). pp. 302–310.
[2] Agrawal, P., Whitaker, R.T., Elhabian, S.Y.: Learning deep features for automated placement of correspondence points on ensembles of complex shapes. Printed in (*). pp. 185–193.
[3] Aksamentov, I., Twinanda, A.P., Mutter, D., Marescaux, J., Padoy, N.: Deep neural networks predict remaining surgery duration from cholecystectomy videos. Printed in (*). pp. 586–593.
[4] Ambrosini, P., Ruijters, D., Niessen, W.J., Moelker, A., van Walsum, T.: Fully automatic and real-time catheter segmentation in x-ray fluoroscopy. Printed in (*). pp. 577–585.
[5] Amit, G., Hadad, O., Alpert, S., Tlusty, T., Gur, Y., BenAri, R., Hashoul, S.: Hybrid mass detection in breast mri combining unsupervised saliency analysis and deep learning. Printed in (*). pp. 594– 602.
[6] Apostolopoulos, S., De Zanet, S., Ciller, C., Wolf, S., Sznitman, R.: Pathological oct retinal layer segmentation using branch residual u-shape networks. Printed in (*).pp. 294–301.
[7] Aubert, B., Vidal, P.A., Parent, S., Cresson, T., Vazquez, C., De Guise, J.: Convolutional neural network and inpainting techniques for the automatic assessment of scoliotic spine surgery from biplanar radiographs. Printed in (*) pp. 691–699.
[8] Badrinarayanan, V., Kendall, A., Cipolla, R.: Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE transactions on pattern analysis and machine intelligence 39(12), 2481–2495 (2017)
[9] Bahrami, K., Rekik, I., Shi, F., Shen, D.: Joint reconstruction and segmentation of 7t-like mr images from 3t mri based on cascaded convolutional neural networks. Printed in (*) pp. 764–772.
[10] Bai, W., Oktay, O., Sinclair, M., Suzuki, H., Rajchl, M., Tarroni, G., Glocker, B., King, A., Matthews, P.M., Rueckert, D.: Semi-supervised learning for network-based cardiac mr image segmentation. Printed in (*). pp. 253–260.
[11] Baur, C., Albarqouni, S., Navab, N.: Semi-supervised deep learning for fully convolutional networks. Printed in (*) pp. 311–319.
[12] Bian, C., Lee, R., Chou, Y.H., Cheng, J.Z.: Boundary regularized convolutional neural network for layer parsing of breast anatomy in automated whole breast ultrasound. Printed in (*). pp. 259–266.
[13] Bortsova, G., van Tulder, G., Dubost, F., Peng, T., Navab, N., van der Lugt, A., Bos, D., De Bruijne, M.: Segmentation of intracranial arterial calcification with deeply supervised residual dropout networks. Printed in (*). pp. 356–364.
[14] Chen, J., Banerjee, S., Grama, A., Scheirer, W.J., Chen, D.Z.: Neuron segmentation using deep complete bipartite networks. Printed in (*). pp. 21–29.
[15] Cheplygina, V., de Bruijne, M., Pluim, J.P.: Not-sosupervised: a survey of semi-supervised, multi-instance, and transfer learning in medical image analysis. arXiv preprint arXiv:1804.06353 (2018)
[16] Dai, L., Sheng, B., Wu, Q., Li, H., Hou, X., Jia, W., Fang, R.: Retinal microaneurysm detection using clinical report guided multi-sieving cnn. Printed in (*). pp. 525–532.
[17] Ding, J., Li, A., Hu, Z., Wang, L.: Accurate pulmonary nodule detection in computed tomography images using deep convolutional neural networks. Printed in (*). pp. 559–567.
[18] Dmitriev, K., Kaufman, A.E., Javed, A.A., Hruban, R.H., Fishman, E.K., Lennon, A.M., Saltz, J.H.: Classification of pancreatic cysts in computed tomography images using a random forest and convolutional neural network ensemble. Printed in (*). pp. 150–158.
[19] Dosovitskiy, A., Fischer, P., Ilg, E., Hausser, P., Hazirbas, C., Golkov, V., van der Smagt, P., Cremers, D., Brox, T.: Flownet: Learning optical flow with convolutional networks. In: Proceedings of the IEEE International Conference on Computer Vision. pp. 2758–2766 (2015)
[20] Dubost, F., Bortsova, G., Adams, H., Ikram, A., Niessen, W.J., Vernooij, M., De Bruijne, M.: Gp-unet: Lesion detection from weak labels with a 3d regression network. Printed in (*). pp. 214–221.
[21] Feng, X., Yang, J., Laine, A.F., Angelini, E.D.: Discriminative localization in cnns for weakly-supervised segmentation of pulmonary nodules. Printed in (*). pp. 568–576.
[22] Fidon, L., Li, W., Garcia-Peraza-Herrera, L.C., Ekanayake, J., Kitchen, N., Ourselin, S., Vercauteren, T.: Scalable multimodal convolutional networks for brain tumour segmentation. Printed in (*). pp. 285–293.
[23] Garg, P., Davenport, E., Murugesan, G., Wagner, B., Whitlow, C., Maldjian, J., Montillo, A.: Using convolutional neural networks to automatically detect eye-blink artifacts in magnetoencephalography without resorting to electrooculography. Printed in (*). pp. 374–381.
[24] Ge, Z., Demyanov, S., Chakravorty, R., Bowling, A., Garnavi, R.: Skin disease recognition using deep saliency features and multimodal learning of dermoscopy and clinical images. Printed in (*). pp. 250–258.
[25] Ghafoorian, M., Mehrtash, A., Kapur, T., Karssemeijer, N., Marchiori, E., Pesteie, M., Guttmann, C.R.G., de Leeuw, F.E., Tempany, C.M., van Ginneken, B., Fedorov, A., Abolmaesumi, P., Platel, B., Wells, W.M.: Transfer learning for domain adaptation in mri: Application in brain lesion segmentation. Printed in (*). pp. 516–524.
[26] Gibson, E., Giganti, F., Hu, Y., Bonmati, E., Bandula, S., Gurusamy, K., Davidson, B.R., Pereira, S.P., Clarkson, M.J., Barratt, D.C.: Towards image-guided pancreas and biliary endoscopy: Automatic multi-organ segmentation on abdominal ct with dense dilated networks. Printed in (*) pp. 728–736.
[27] Gupta, V., Thomopoulos, S.I., Rashid, F.M., Thompson, P.M.: Fibernet: An ensemble deep learning framework for clustering white matter fibers. Printed in (*). pp. 548–555.
[28] Hochreiter, S., Schmidhuber, J.: Long short-term memory. Neural Computation 9(8), 1735–1780 (1997), https://doi.org/10.1162/neco.1997.9.8.1735
[29] Hou, B., Alansary, A., McDonagh, S., Davidson, A., Rutherford, M., Hajnal, J.V., Rueckert, D., Glocker, B., Kainz, B.: Predicting slice-to-volume transformation in presence of arbitrary subject motion. Printed in (*). pp. 296–304.
[30] Huang, W., Bridge, C.P., Noble, J.A., Zisserman, A.: Temporal heartnet: Towards human-level automatic analysis of fetal cardiac screening video. Printed in (*). pp. 341–349.
[31] Iglesias, J.E., Lerma-Usabiaga, G., Garcia-Peraza-Herrera, L.C., Martinez, S., Paz-Alonso, P.M.: Retrospective head motion estimation in structural brain mri with 3d cnns. Printed in (*). pp. 314–322.
[32] Jaderberg, M., Simonyan, K., Zisserman, A., kavukcuoglu, k.: Spatial transformer networks. In: Cortes, C., Lawrence, N.D., Lee, D.D., Sugiyama, M., Garnett, R. (eds.) Advances in Neural Information Processing Systems 28, pp. 2017–2025. Curran Associates, Inc. (2015)
[33] Johansen-Berg, H., Behrens, T.E.: Diffusion MRI: from quantitative measurement to in vivo neuroanatomy. Academic Press (2013)
[34] Joyce, T., Chartsias, A., Tsaftaris, S.A.: Robust multi-modal mr image synthesis. Printed in (*). pp. 347–355.
[35] Karani, N., Tanner, C., Kozerke, S., Konukoglu, E.: Temporal interpolation of abdominal mris acquired during freebreathing. Printed in (*). pp. 359–367.
[36] Karimi, D., Ruan, D.: Synergistic combination of learned and hand-crafted features for prostate lesion classification in multiparametric magnetic resonance imaging. Printed in (*). pp. 391–398.
[37] Kingma, D.P., Salimans, T., Welling, M.: Variational dropout and the local reparameterization trick. In: Cortes, C., Lawrence, N.D., Lee, D.D., Sugiyama, M., Garnett, R. (eds.) Advances in Neural Information Processing Systems 28, pp. 2575–2583. Curran Associates, Inc. (2015), http://papers.nips.cc/paper/5666-variational-dropout-and-the-local-reparameterization-trick.pdf
[38] Kiraly, A.P., Nader, C.A., Tuysuzoglu, A., Grimm, R., Kiefer, B., El-Zehiry, N., Kamen, A.: Deep convolutional encoderdecoders for prostate cancer detection and classification. Printed in (*). pp. 489–497.
[39] Ktena, S.I., Parisot, S., Ferrante, E., Rajchl, M., Lee, M., Glocker, B., Rueckert, D.: Distance metric learning using graph convolutional networks: Application to functional brain networks. Printed in (*) pp. 469–477.
[40] Kurmann, T., Marquez Neila, P., Du, X., Fua, P., Stoyanov, D., Wolf, S., Sznitman, R.: Simultaneous recognition and pose estimation of instruments in minimally invasive surgery. Printed in (*). pp. 505– 513.
[41] Lafferty, J., McCallum, A., Pereira, F.C.: Conditional random fields: Probabilistic models for segmenting and labeling sequence data (2001)
[42] Laina, I., Rieke, N., Rupprecht, C., Vizca´ıno, J.P., Eslami, A., Tombari, F., Navab, N.: Concurrent segmentation and localization for tracking of surgical instruments. Printed in (*). pp. 664–672.
[43] Litjens, G., Kooi, T., Bejnordi, B.E., Setio, A.A.A., Ciompi, F., Ghafoorian, M., van der Laak, J.A., van Ginneken, B., S´anchez, C.I.: A survey on deep learning in medical image analysis. Medical image analysis 42, 60–88 (2017)
[44] Liu, M., Zhang, J., Adeli, E., Shen, D.: Deep multi-task multi-channel learning for joint classification and regression of brain status. Printed in (*). pp. 3–11.
[45] Meng, Q., Roth, H.R., Kitasaka, T., Oda, M., Ueno, J., Mori, K.: Tracking and segmentation of the airways in chest ct using a fully convolutional network. Printed in (*).pp. 198–207.
[46] Milletari, F., Rothberg, A., Jia, J., Sofka, M.: Integrating statistical prior knowledge into convolutional neural networks. Printed in (*) pp. 161–168.
[47] Nardelli, P., Ross, J.C., Est´epar, R.S.J.: Ct image enhancement for feature detection and localization. Printed in (*). pp. 224–232.
[48] Oktay, O., Ferrante, E., Kamnitsas, K., Heinrich, M., Bai, W., Caballero, J., Cook, S.A., de Marvao, A., Dawes, T., ORegan, D.P., et al.: Anatomically constrained neural networks (acnns): application to cardiac image enhancement and segmentation. IEEE transactions on medical imaging 37(2), 384–395 (2018)
[49] Parajuli, N., Lu, A., Stendahl, J.C., Zontak, M., Boutagy, N., Alkhalil, I., Eberle, M., Lin, B.A., O’Donnell, M., Sinusas, A.J., Duncan, J.S.: Flow network based cardiac motion tracking leveraging learned feature matching. Printed in (*) pp. 279–286.
[50] Poulin, P., Côté, M.A., Houde, J.C., Petit, L., Neher, P.F., Maier-Hein, K.H., Larochelle, H., Descoteaux, M.: Learn to track: Deep learning for tractography. Printed in (*) pp. 540–547.
[51] Pourtaherian, A., Ghazvinian Zanjani, F., Zinger, S., Mihajlovic, N., Ng, G., Korsten, H., de With, P.: Improving needle detection in 3d ultrasound using orthogonal-plane convolutional networks. Printed in (*). pp. 610–618.
[52] Ravishankar, H., Venkataramani, R., Thiruvenkadam, S., Sudhakar, P., Vaidya, V.: Learning and incorporating shape models for semantic segmentation. Printed in (*) pp. 203–211.
[53] Ronneberger, O., Fischer, P., Brox, T.: U-net: Convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. pp. 234–241. Springer International Publishing, Cham (2015)
[54] Roy, A.G., Conjeti, S., Sheet, D., Katouzian, A., Navab, N., Wachinger, C.: Error corrective boosting for learning fully convolutional networks with limited data. Printed in (*). pp. 231–239.
[55] Salehi, M., Prevost, R., Moctezuma, J.L., Navab, N., Wein, W.: Precise ultrasound bone registration with learningbased segmentation and speed of sound calibration. Printed in (*). pp. 682–690.
[56] Sedai, S., Mahapatra, D., Hewavitharanage, S., Maetschke, S., Garnavi, R.: Semi-supervised segmentation of optic cup in retinal fundus images using variational autoencoder. Printed in (*). pp. 75–82.
[57] Shen, H., Wang, R., Zhang, J., McKenna, S.J.: Boundaryaware fully convolutional network for brain tumor segmentation. Printed in (*). pp. 433–441.
[58] Sinclair, M., Bai, W., Puyol-Antón, E., Oktay, O., Rueckert, D., King, A.P.: Fully automated segmentation-based respiratory motion correction of multiplanar cardiac magnetic resonance images for large-scale datasets. Printed in (*) pp. 332–340.
[59] Sokooti, H., de Vos, B., Berendsen, F., Lelieveldt, B.P.F., Isgum, I., Staring, M.: Nonrigid image registration using multi-scale 3d convolutional neural networks. Printed in (*) pp. 232–239.
[60] Tanno, R., Ghosh, A., Grussu, F., Kaden, E., Criminisi, A., Alexander, D.C.: Bayesian image quality transfer. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. pp. 265–273. Springer (2016)
[61] Tanno, R., Worrall, D.E., Ghosh, A., Kaden, E., Sotiropoulos, S.N., Criminisi, A., Alexander, D.C.: Bayesian image quality transfer with cnns: Exploring uncertainty in dmri super-resolution. Printed in (*) pp. 611–619.
[62] Uzunova, H., Wilms, M., Handels, H., Ehrhardt, J.: Training cnns for image registration from few samples with modelbased data augmentation. Printed in (*) pp. 223–231.
[63] Wang, Z., Yin, Y., Shi, J., Fang, W., Li, H., Wang, X.: Zoomin-net: Deep mining lesions for diabetic retinopathy detection. Printed in (*). pp. 267–275.
[64] Xie, Y., Xia, Y., Zhang, J., Feng, D.D., Fulham, M., Cai, W.: Transferable multi-model ensemble for benign-malignant lung nodule classification on chest ct. Printed in (*).pp. 656–664.
[65] Xu, C., Xu, L., Gao, Z., Zhao, S., Zhang, H., Zhang, Y., Du, X., Zhao, S., Ghista, D., Li, S.: Direct detection of pixel-level myocardial infarction areas via a deep-learning algorithm. Printed in (*). pp. 240–249.
[66] Yang, D., Xiong, T., Xu, D., Zhou, S.K., Xu, Z., Chen, M., Park, J., Grbic, S., Tran, T.D., Chin, S.P., Metaxas, D., Comaniciu, D.: Deep image-to-image recurrent network with shape basis learning for automatic vertebra labeling in largescale 3d ct volumes. Printed in (*). pp. 498–506.
[67] Yang, X., Yu, L., Li, S., Wang, X., Wang, N., Qin, J., Ni, D., Heng, P.A.: Towards automatic semantic segmentation in volumetric ultrasound. Printed in (*) pp. 711–719.
[68] Yang, Y., Li, T., Li, W., Wu, H., Fan, W., Zhang, W.: Lesion detection and grading of diabetic retinopathy via two-stages deep convolutional neural networks. Printed in (*) pp. 533–540.
[69] Yao, J., Kovacs, W., Hsieh, N., Liu, C.Y., Summers, R.M.: Holistic segmentation of intermuscular adipose tissues on thigh mri. Printed in (*) pp. 737–745.
[70] Ye, C.: Learning-based ensemble average propagator estimation. Printed in (*) pp. 593–601.
[71] Ye, C., Prince, J.L.: Fiber orientation estimation guided by a deep network pp. 575–583 (2017)
[72] Yu, L., Cheng, J.Z., Dou, Q., Yang, X., Chen, H., Qin, J., Heng, P.A.: Automatic 3d cardiovascular mr segmentation with densely-connected volumetric convnets. Printed in (*). pp. 287–295.
[73] Zhang, J., Liu, M., Wang, L., Chen, S., Yuan, P., Li, J., Shen, S.G.F., Tang, Z., Chen, K.C., Xia, J.J., Shen, D.: Joint craniomaxillofacial bone segmentation and landmark digitization by context-guided fully convolutional networks. Printed in (*). pp. 720–728.
[74] Zheng, J., Miao, S., Liao, R.: Learning cnns with pairwise domain adaption for real-time 6dof ultrasound transducer detection and tracking from x-ray images. Printed in (*) pp. 646–654.
[75] Zhou, Y., Xie, L., Fishman, E.K., Yuille, A.L.: Deep supervision for pancreatic cyst segmentation in abdominal ct scans. Printed in (*). pp. 222–230.
[76] Zhu, W., Lou, Q., Vang, Y.S., Xie, X.: Deep multi-instance networks with sparse label assignment for whole mammogram classification. Printed in (*). pp. 603–611.