In humans, each cell generally contains 23 pairs of chromosomes (totaling 46 chromosomes): 22 pairs are called autosomes and look the same in females and males. The 23rd pair (the two sex chromosomes) differ between females and males in that females have two X chromosomes and males have one X and one Y chromosome.
Several features help identify the chromosome within the individual karyotype set: the distribution of gray shades along the medial axis, the position of the centromere (a limited region of the chromosome, which might be challenging to identify) and the relative length of the chromosome in the set.
Cytologists able to perform a manual chromosome classification are rare and the results they obtain could be improved in terms of time, and costs when supported or replaced by machine learning systems. Most of the research activity in computer vision in recent years show very high capabilities on different types of datasets and applications and RSIP Vision has developed much experience in the machine learning field.
Chromosomes classification with deep learning
We decided to develop a solution to our client’s need by means of convolutional neural networks (CNN), a deep learning technique currently deployed in many practical applications like image recognition, speech recognition, photo taggers and many more. CNN are extremely valuable when nearby values of array data are correlated. Ideally, the convolutional neural networks should be trained on a very large dataset. It can also be trained on one system with sufficient number of cases and then fine-tuned to work on a new system.
We used the convolutional neural networks in first place as feature extractor; then we aligned and centered the data; finally, we extracted the features map from each image and trained them on a linear 1d classifier.
Our client received a very robust solution for chromosome classification, which will keep becoming more and more solid as it is given additional datasets to train on.

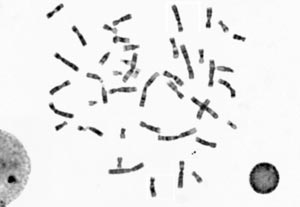
 Microscopy
Microscopy