Seed Detection
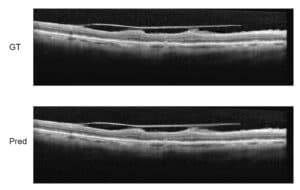
We’ve been working on automatically detecting Cystoid Macular Edema (CME) shown in Optical Coherence Tomography (OCT) images. CME is a buildup of fluids in the eye near the macula of the eye. OCT is one of the main modalities used to diagnose CME.
Our goal is to develop a system that can process large amounts of OCT images, automatically singling out images that show CME and quantify the cysts.
We have developed a multi-process method for more efficient automatic detection. The previous articles described the overall detection method, the denoising process, and layer segmentation. The next step is seed detection.
The goal of seed detection is to identify super-pixels in the scans that belong to a cyst. To do this, we will use machine learning methods with robust image processing features and build a classifier that can automatically detect cyst super-pixels in new images.
Super Pixels
Super-pixels are an increasingly popular computer vision concept where neighboring pixels are grouped into coherent units with perceptually shared meanings. Using this concept, a super-pixel carries much more meaning both geometrically and texturally than a single pixel. This allows us to create a meaningful classifier for super-pixels.
Our classifier will take into consideration the textural features of the super-pixel as well as its spatial location (which correlates to the distance of the area from the fovea) to determine whether the super-pixel belongs to a cyst or not.
As our features we use both statistics (skewness, kurtosis and more), spatial features (x,y coordinates) and various textural features (most prominent are the gabor filters, local binary patterns (LBP) and acutance).
Our Process
First we pre-process the image by cropping out the retina using the previously segmented RPE and ILM (see Layer Segmentation section for more detail). Next we segment the retina into super-pixels using the SLIC algorithm, which was introduced in the Denoising process.
From each super-pixel we extract the features mentioned before and train an ensemble of random forests combined with the adaBoost method to select the best features and cross-validate the results using the leave-one-out approach.
Estimating our Results
We had two manual annotations of the scans and due to the difficulty and subjectivity of defining a cyst have chosen to take a conservative approach and consider a proper “cyst” only cysts that were detected in both annotations.
In general we have found that using an ensemble of 32 forests is optimal with an average of 0.9243 accuracy, 0.92 sensitivity and 0.9253 specificity.
We expect that by adding data from neighboring super-pixels to our classifier we will be able to get much better results.

 Ophthalmology
Ophthalmology