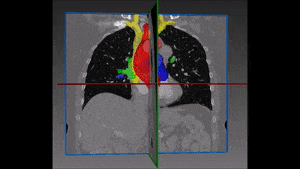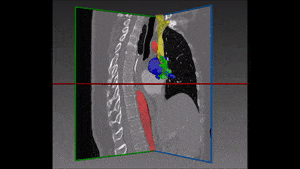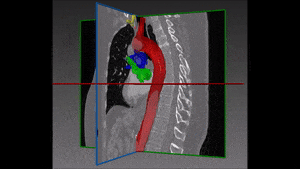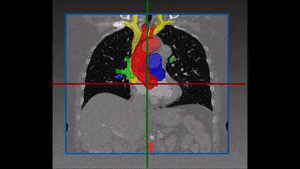
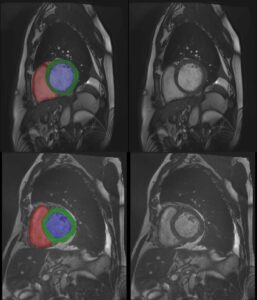
1.1 Segmentation tasks
[10] suggest a new fully convolutional network architecture for the task of cardiovascular MRI segmentation. The architecture is based on the idea of network blocks in which each layer is densely connected with auxiliary side paths (skip connections) to all the following layers in the block. The main advantage in the structure is the reduced number of parameters, which makes training and prediction more efficient than standard CNNs. Parameter reduction is especially important in volumetric data, where the number of voxels scale cubically with the resolution and the size of the network may make the difference between and applicable and non-applicable solution. The proposed model improved over the results in the HVSMR 2016 challenge for whole-heart and great vessel segmentation from 3D cardiac MRI, while about ten times less parameters than U-Net, and about half the number of parameters than other competing networks. The Dice scores on the HVSMR 2016 data are 82% and 93% on myocardium and blood pool segmentation, respectively.
A different approach for reducing the number of parameters is presented in [5] for segmentation of the left atrium and proximal pulmonary veins from MRI data. Instead of taking the whole MRI volume as input, they use three orthogonal projections of the volume as inputs to three CNNs, which are fused together to one network. Another interesting technical detail in this model is the uncommon use of the z-loss [3], which the authors report leads to a faster convergence of the training process. The method achieved a Dice score of 90% on the STACOM 2013 challenge data, improving a little above 1% over previous state-of-the-art result.
Other ways suggested for enhancing segmentation performance involve combination of additional data together with convolutional networks. [4] show how to integrate prior statistical knowledge, obtained through principal components analysis (PCA), into a convolutional neural network in order to obtain segmentation predictions with corrupted or noisy cardiac ultrasound images. They show how to perform two different tasks: segmentation of the left ventricle of the heart from apical-view ultrasound scans, and landmark localization for 14 points of interest in images from the parasternal-long-axis view. Their network was trained and evaluated on datasets of total sizes of 1,100 images for the segmentation task, and 800 images for the landmark localization task, and achieved on average similar Dice score to U-Net, but with a more stable performance: on average the U-Net Dice score was 88%, but ranged from 63% to 96% on different examples, while the scores of the proposed model were on average 87% and ranged between 80% and 96%.
[2] propose a semi-supervised learning approach, in which a segmentation CNN is trained from both labeled and unlabeled data. At the first stage, initial training is performed only on the labeled dataset, and at the second training stage the network parameters and the segmentations for the unlabeled data are alternately updated. They evaluate the method for cardiac MRI segmentation on a dataset of 100 MRI scans from the UK Biobank study. In this model, segmentation is performed on each MRI slice separately, and the model achieves Dice scores of 92%, 85% and 89% on segmentation of the left ventricular cavity, the myocardium and the right-ventricle, respectively.
1.2 Temporal monitoring
Analysis of sequential cardiac ultrasound echocardiography scans (echocine) is a task that highly depends on the expertise of the clinicians analyzing the data and has a high variance across different experts. Recent works suggest deep learning tools for motion tracking [6] and for automated evaluation of cine echo measurements [1]. [6] use deep learning in a non-direct manner for cardiac motion tracking. They model the motion as a flow in a graph, in which the nodes represent different image patches. The flow is calculated with linear programing under a model of physiological constraints and conservation laws of the ingoing and outgoing flows at each site. The authors show that it is possible to use a Siamese CNN to learn similarity and dissimilarity between pairs of image patches (represented in the graph as nodes) to generate the edge weights of a weighted graph. The model is evaluated in the paper on simulated data and shows promising results.
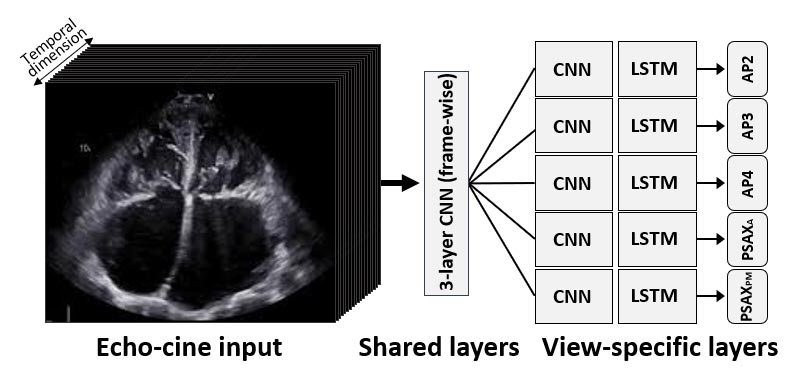
[1] design a new network architecture, based on five streams of Convolution+LSTM networks for automatic evaluation of quality scores to echo sequences. The network (see figure below) starts with shared convolution and max-pooling layers which input the 2D images frame-by-frame and learn low-level features of the images; and then the output sequence of this CNN splits to five streams of convolution and LSTM layers. Each stream is trained on a different 2D view plane of the ultrasound scan. The LSTM layers are used here as integrators of the temporal relations between the frames in the sequence, and generate a single output for each sequence. At the training stage the network is trained on regression with l2 regularization and l1 loss in respect to gold-standard scores. An evaluation of this model on 2,450 echo cines of 509 patients gave between 83% and 89% accuracy on sequences from the five views.
[9] use deep learning in cardiology to perform detection of myocardial infarction (MI) from cardiac MRI. Their network combines optical flow, convolutional layers, LSTM and then fully connected layers, and performs classification at the pixel level to segment the MI regions. The authors report accuracy of 94.3% and precision of 91.3% on a dataset of 114 subjects.
1.3 Registration with motion correction
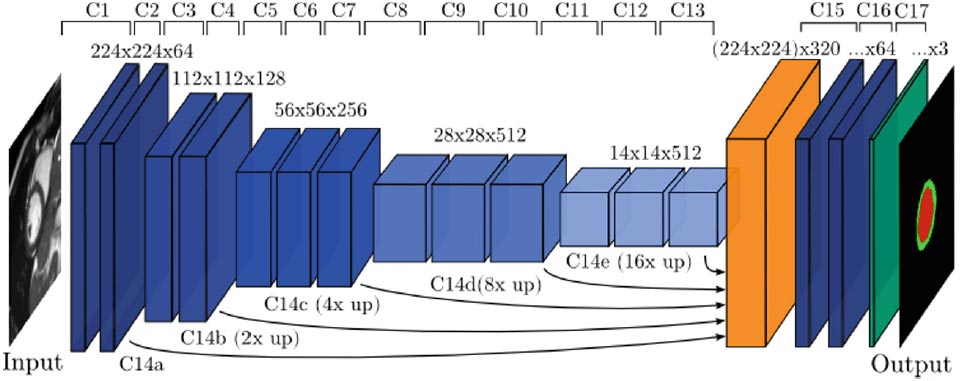
Cardiac magnetic resonance (CMR) slice to volume registration requires alignment of 2D images that are often acquired at different breath-holds. The CMR slices are taken at synchronized cardiac phases, as measured by EEG; however, the body motion between slices creates non-rigid transformations – making accurate registration a hard problem, especially 2 for accurate alignment of the myocardium. [8] show how to use a CNN to correct the motion and perform the between-slice registration to a 3D volume. They use a VGG-like architecture (see figure below) with 17 layers and achieve Dice scores around 88% on a CMR dataset from the UK Biobank database.
.
Deep Learning in Cardiology – References
(*) Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) Medical Image Computing and Computer-Assisted Intervention MICCAI 2017. Springer International Publishing, Cham (2017)
[1] Abdi, A.H., Luong, C., Tsang, T., Jue, J., Gin, K., Yeung, D., Hawley, D., Rohling, R., Abolmaesumi, P.: Quality assessment of echocardiographic cine using recurrent neural networks: Feasibility on five standard view planes. Printed in (*) pp. 302–310
[2] Bai, W., Oktay, O., Sinclair, M., Suzuki, H., Rajchl, M., Tarroni, G., Glocker, B., King, A., Matthews, P.M., Rueckert, D.: Semi-supervised learning for network-based cardiac mr image segmentation. Printed in (*) pp. 253–260.
[3] de Br´ebisson, A., Vincent, P.: The z-loss: a shift and scale invariant classification loss belonging to the spherical family. arXiv preprint arXiv:1604.08859 (2016)
[4] Milletari, F., Rothberg, A., Jia, J., Sofka, M.: Integrating statistical prior knowledge into convolutional neural networks. Printed in (*) pp. 161–168.
[5] Mortazi, A., Karim, R., Rhode, K., Burt, J., Bagci, U.: Cardiacnet: Segmentation of left atrium and proximal pulmonary veins from mri using multi-view cnn Printed in (*) pp. 377–385.
[6] Parajuli, N., Lu, A., Stendahl, J.C., Zontak, M., Boutagy, N., Alkhalil, I., Eberle, M., Lin, B.A., O’Donnell, M., Sinusas, A.J., Duncan, J.S.: Flow network based cardiac motion tracking leveraging learned feature matching. Printed in (*) pp. 279–286.
[7] Simonyan, K., Zisserman, A.: Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556 (2014)
[8] Sinclair, M., Bai, W., Puyol-Ant´on, E., Oktay, O., Rueckert, D., King, A.P.: Fully automated segmentation-based respiratory motion correction of multiplanar cardiac magnetic resonance images for large-scale datasets. Printed in (*) pp. 332–340.
[9] Xu, C., Xu, L., Gao, Z., Zhao, S., Zhang, H., Zhang, Y., Du, X., Zhao, S., Ghista, D., Li, S.: Direct detection of pixel-level myocardial infarction areas via a deep-learning algorithm. Printed in (*) pp. 240–249.
[10] Yu, L., Cheng, J.Z., Dou, Q., Yang, X., Chen, H., Qin, J., Heng, P.A.: Automatic 3d cardiovascular mr segmentation with densely-connected volumetric convnets. Printed in (*) pp. 287–295.

 Cardiology
Cardiology